HIBLUP can construct XRM from pedigree and genomic information, the output binary file has two types of storage format. For dense matrix, the values at lower triangle of relationship matrix are stored one by one, one element take over 4 bytes, and for sparse matrix, only non-zero values (k) at lower triangle of relationship matrix and its row (i) and column index (j) are stored in order of “ijk” format in binary file, that means one element will take over 12 bytes.
Pedigree based relationship matrix (PRM)
# construct A and its inverse simultaneously
./hiblup --make-xrm
--pedigree demo.ped
--add
--add-inv #wether to construct its inverse matrix
--thread 32
--out demo
Files named demo.PA.bin, demo.PA.id, demo.PAinv.sparse.bin, demo.PAinv.sparse.id will be generated in the work directory.
Since the binary file (*.bin) cannot be opened directly, users can add a flag --write-txt in the commands to output a readable “.txt” file.
Sometimes when computing the inverse of XRM, it may encounter an error saying:
Error: matrix is not invertible,…
that is because that the matrix is not positive-defined, users can specify a small value to --ridge-value by using ridge regression method to address this problem.
Genome based relationship matrix (GRM)
The relationship matrix could also be derived from genome-wide markers. The mathematical formula of constructing a genomic relationship matrix (GRM) is as follows:
\begin{equation}
\begin{aligned}
G &= \frac{ZZ^\top}{tr(ZZ^\top)/n}
\end{aligned}
\end{equation}where Z is the numeric coded genotype matrix with a dimension of n by m, n is the number of individuals and m is the number of markers, tr is the the trace of the matrix. Here in HIBLUP, total 4 methods have been achieved to obtain matrix Z for additive and dominant genetic effect, the way of coding the genotype for different methods can be referred to the following table:
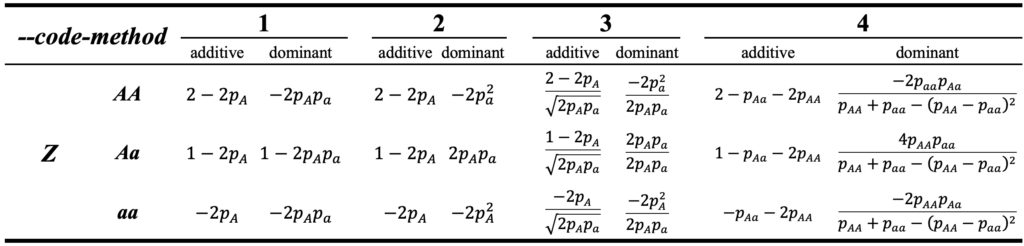
where p_{A} and p_{a} are the allele frequency of A and a, respectively. p_{AA}, p_{Aa} and p_{aa} are the genotype frequency of AA, Aa, aa, respectively.
Users can easily switch to any of method above by specifying the flag --code-method with corresponding value, e.g. --code-method 2. The reference for different methods can be found at the following papers:
--code-method 1, (default) Su, Guosheng, et al. “Estimating additive and non-additive genetic variances and predicting genetic merits using genome-wide dense single nucleotide polymorphism markers.” Plos one (2012): e45293.--code-method 2, Zeng, Zhao-Bang, Tao Wang, and Wei Zou. “Modeling quantitative trait loci and interpretation of models.” Genetics 169.3 (2005): 1711-1725.--code-method 3, Yang, Jian, et al. “Common SNPs explain a large proportion of the heritability for human height.” Nature genetics 42.7 (2010): 565-569.--code-method 4, Vitezica, Zulma G., et al. “Orthogonal estimates of variances for additive, dominance, and epistatic effects in populations.” Genetics 206.3 (2017): 1297-1307.
General GRM
The script to construct additive and dominant genomic relationship matrix:
# construct A and D simultaneously
./hiblup --make-xrm --code-method 2
--bfile demo
--add --dom
--step 10000
--thread 32
--out demo
#--write-txt #to output a readable ".txt" file
Files named demo.GA.bin, demo.GA.id, demo.GD.bin, demo.GD.id will be generated in the work directory, and HIBLUP will directly compute the inverse of the GRM and output it if --add-inv or --dom-inv is detected in the command.
Note that users can assign a file listing the names of SNPs to the flag --extract or --exclude to construct GRM by a subset of SNPs. For example, constructing GRM only uses the significant SNPs:
./hiblup --make-xrm
--bfile demo
--add
--extract sig.snp.txt
--thread 32
--out demo
where the file sig.snp.txt contains the SNPs which are considered to be significantly associated with a trait.
Mixed GRM for multiple populations
HIBLUP can construct a mixed GRM if the genotype data has multiple populations (e.g., different breeds), the mathematical formula of mixed GRM can be expressed as follows:
\begin{equation}
\begin{aligned}
G = \left[\begin{matrix}G_{ii}&G_{ij}\\G_{ji}&G_{jj}\\\end{matrix}\right]=\left[\begin{matrix}\frac{Z_{i}Z_{i}^\top}{tr(Z_{i}Z_{i}^\top)/n_{i}}&\frac{Z_{i}Z_{j}^\top}{\sqrt{tr(Z_{i}Z_{i}^\top)\ast tr(Z_{j}Z_{j}^\top)/(n_{i}\ast n_{j})}}\\\frac{Z_{j}Z_{i}^\top}{\sqrt{tr(Z_{j}Z_{j}^\top)\ast tr(Z_{i}Z_{i}^\top)/(n_{j}\ast n_{i})}}&\frac{Z_{j}Z_{j}^\top}{tr(Z_{j}Z_{j}^\top)/n_{j}}\\\end{matrix}\right]
\end{aligned}
\end{equation}where Z_{i} and Z_{j} are the centered or scaled genotype matrix (as shown in above Table 1) using corresponding allele frequency or genotype frequency of i_{th} and j_{th} population, respectively. n_{i} and n_{j} are the sample size of different populations.
It’s pretty easy to implement by just providing a population classification file (see file format here) when running HIBLUP to construct a regular GRM:
# construct mixed GRM
./hiblup --make-xrm --code-method 1
--bfile demo
--pop-class demo.popclass.txt
--step 10000
--thread 32
--out demo
Weighted GRM
HIBLUP supports to compute the weighted GRM by the following script:
./hiblup --make-xrm --code-method 1
--bfile demo
--add --dom
--snp-weight snp.wt.txt
--thread 32
--out demo
The file format of SNP weights please refer to here.
Pedigree and Genome based relationship matrix (HRM)
When pedigree and genome information are provided simultaneously, HIBLUP will automatically construct H matrix following the equation below (Aguilar 2010, Christensen 2010):
\begin{equation}
H =
\begin{bmatrix}
A_{11} + A_{12} A_{22}^{-1} (G_w - A_{22}) A_{22}^{-1} A_{21} & A_{12}A_{22}^{-1}G_w \\
G_wA_{22}^{-1}A_{21} & G_w \\
\end{bmatrix}
\end{equation}where A_{11} and A_{22} are the submatrices of A matrix for non-genotyped and genotyped individuals, respectively; A_{12} and A_{21} are the submatrices of A matrix, which represent the relationships between genotyped and nongenotyped individuals, G_w = (1 - alpha) * G_{adj} + alpha * A_{22}, G_{adj} is adjusted from the original genomic relastionship matrix G by simple linear regression:
\begin{equation}
\begin{aligned}
Avg(diag(G)) * a + b = Avg(diag(A_{22})) \\
Avg(offdiag(G)) * a + b = Avg(offdiag(A_{22}))
\end{aligned}
\end{equation}Then we have G_{adj} = G * a + b.
./hiblup --make-xrm
--pedigree demo.ped
--bfile demo
--alpha 0.05
--add --dom
--out demo
The above command will generate additive and dominant relationship matrix simultaneously, the filenames are: demo.HA.bin, demo.HA.id, demo.HD.bin, demo.HD.id.
Users can also specify --snp-weight to construct a weighted GRM or specify --pop-class to construct a mixed GRM for constructing HRM.
Besides providing binary genotype to construct HRM, users can also use their own pre-computed GRM to construct HRM, but the pre-computed GRM should be stored in binary format, which can be transformed from square or triangle text file by HIBLUP (see Format conversion of XRM), the corresponding commands are as follows:
./hiblup --make-xrm
--pedigree demo.ped
--xrm yourGRM # yourGRM.bin, yourGRM.id
--alpha 0.05
--add # or '--dom' is supported
--out demo
Also, HIBLUP can construct the inverse of H matrix by following script:
./hiblup --make-xrm
--pedigree demo.ped
--bfile demo
--alpha 0.05
--add-inv --dom-inv
--out demo
the generated filenames are: demo.HAinv.sparse.bin, demo.HAinv.sparse.id, demo.HDinv.sparse.bin, demo.HD.sparse.id.
As described above, you can also construct the inverse of HRM using your own pre-computed GRM.
XRM for environmental random effect
Except for constructing XRM for genetic random effect, HIBLUP can also construct XRM for environmental random effect, phenotype file should be provided for this case, the command is as following:
./hiblup --make-xrm
--pheno demo.phe
--rand 6,7
--out demo
the generated filenames are: demo.loc.sparse.bin, demo.lco.sparse.id, demo.dam.sparse.bin, demo.dam.sparse.id.
NOTE
(1) All above relationship matrix, except for those whose prefix contain character “.sparse.”, can be used by GCTA, LDAK, but additional processes are needed, for example:
awk '{print $1,$1}' demo.GA.id > gcta.grm.id
cp demo.GA.bin gcta.grm.bin
# then use it by GCTA
./gcta64 ... --grm gcta ...
(2) The binary format relationship matrix calculated by GCTA or LDAK can be used directly by HIBLUP without any format transformation, just assign its prefix to flag --xrm.